Messing Lab
Research Overview
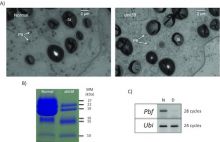
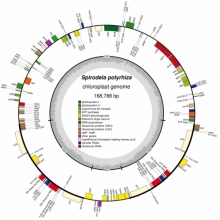
The Messing lab would like to contribute to the understanding of the expression of regulation of gene copies in plants. It is now apparent that many gene products are derived from multiple gene copies. As use of copies rapidly increases, their sequences are quite conserved or so similar that it becomes difficult to infer gene products from which the genes they produce. Therefore, it becomes necessary to sequence the genome of an organism so that one can sort gene copies in their location on chromosomes. Then one can match each RNA species quantitatively with individual gene copies.
Contact Information
Waksman Institute
190 Frelinghuysen Road
Messing Lab
Piscataway, NJ 08854
United States
Links
Selected Publications
Complete list of publications: [Pubmed]
Alumni
Former Postdoctoral Fellows
- Dr. Zhiyong Zhang
- Chenxu Liu
Former Lab Researchers
- Jennifer Ayer