Gene expression and evolution of seed proteins
An important aspect of gene expression is DNA modification and chromatin structure. Maize seems to be in particular suited for this purpose because the portion of the genome representing active genes is rather small. The maize genome has an even higher percentage of repeat elements than the human genome, 85% versus 50%. Therefore, detection of such epigenetic marks in maize had to be highly enriched for a fraction of the genome. Indeed, this specific histone acetylation correlated well with transcribed genes and could be verified with many known examples of genetic loci.
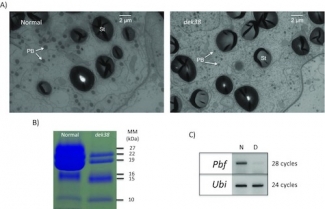
Transcriptional analysis of seed development has shown that many of them are not expressed despite a normal gene structure. That has prompted us to investigate how epigenetic silencing could play a role in repressing gene expression. We applied bisulfite DNA sequencing, which can detect the methylation of DNA sequences. The methylation pattern of specific gene clusters can then be related to the transcriptional activity of them during seed development. There were several findings:
- Genes have a higher degree of methylation in non-expressing tissues, which is consistent with the observation that active genes are hypomethylated.
- The difference in methylation, however, can differ between chromosomal locations.
- Culturing endosperm, which keeps cells dividing, not only reduces DNA methylation, but also can lead to transcription of genes that were previously inactive