A pathway for synapsis initiation during zygotene in Drosophila oocytes
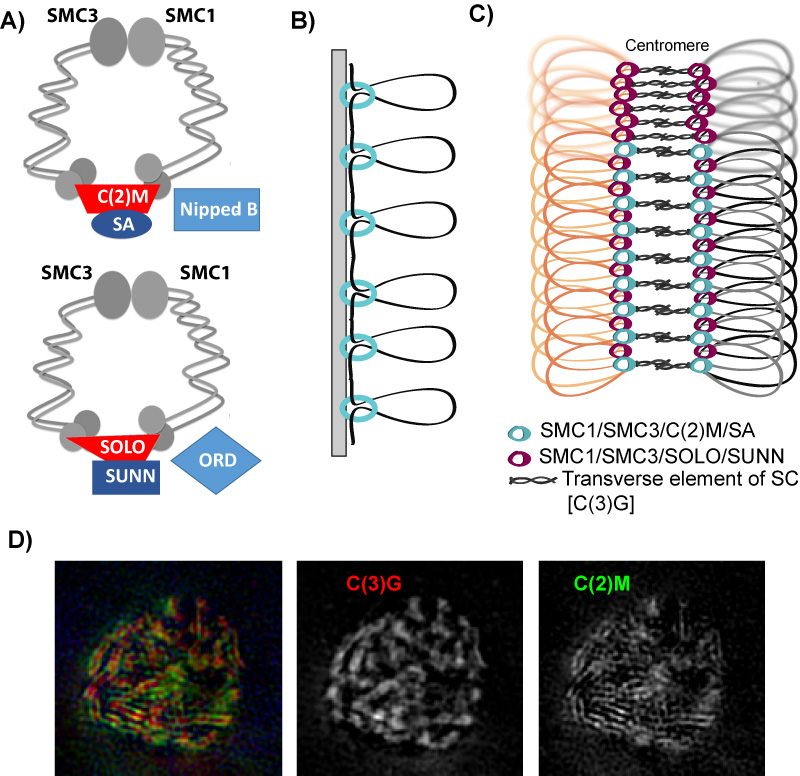
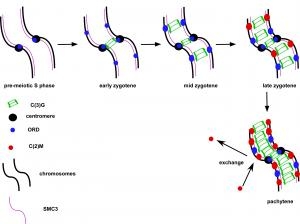
Pairs of meiotic chromosomes, or homologs, are brought together in an elaborate pairing process which culminates with synapsis, where bivalents are held together along their entire length by the synaptonemal complex (SC). Formation of the SC between homologs is essential for crossing over and chromosome segregation but how the homologs are paired and SC assembly initiates is poorly understood. We investigated the requirements for SC assembly in Drosophila oocytes and found that there are three temporally and genetically distinct stages of synapsis initiation (Figure 1). First, early zygotene cells have one or two patches of SC that colocalizes with a cluster of centromeres, suggesting synapsis initiates first at the centromeres. Second, oocytes at mid-zygotene contain the centromere SC plus several euchromatic sites. The centromeric and first euchromatic SC initiation sites depend on the cohesin protein ORD. Third, late zygotene contains many more sites of SC initiation and these depend on the Kleisin-like protein C(2)M. Surprisingly, the synapsis initiation events in late zygotene are independent of the earlier mid-zygotene events but all synapsis depend on the cohesin subunit SMC3. Our results show that cohesin proteins have an important role in SC initiation. Based on the observation that ORD and SMC3 are enriched at centromeres and promote their clustering and synapsis, we suggest that the enrichment of cohesin proteins at specific sites promotes homolog interactions and the initiation of euchromatic SC assembly. ORD is also required for most crossing over, suggesting that SC initiation at mid-zygotene may also trigger the formation of DSBs that will be repaired as crossovers.
Page Components