Plastid Transformation in Arabidopsis
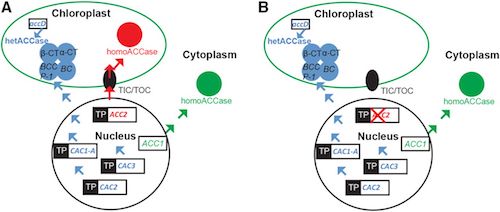
Plastid transformation has been inefficient in Arabidopsis thaliana due to a natural tolerance of Arabidopsis to spectinomycin, the selective agent employed to enrich transformed plastid genomes. Tolerance to spectinomycin has been linked to a duplication of the ACCase biosynthetic pathway in chloroplasts. We have shown that plastid transformation is 100-fold more efficient in Arabidopsis lines defective in the plastid-targeted ACC2 nuclear gene (Yu et al. Plant Physiol. 175: 186-193, 2017). This information has been obtained in the the Col-0 ecotype that is recalcitrant to plant regeneration. We now report ACC2 defective lines in the RLD and Ws ecotypes, which readily regenerate plants from cultured cells. ACC2 knockouts were obtained using CRISPR/Cas9 genome editing tools. The spectinomycin hypersensitive phenotype is characterized by the lack of shoot apex when germinated on a selective medium. This phenotype has been confirmed in both accessions, indicating that deletion of the ACC2 gene is generally applicable to obtain spectinomycin hypersensitive plants in all species in which duplication of the ACCase pathway has been conserved. Testing plastid transformation efficiency in the ACC2 knockout lines confirmed 100-fold elevated frequency as compared to the wild-type. Testing was accelerated by the newly developed SPEED transformation protocol (Yu et al., Plant Physiol. 181: 394-398, 2020). The research is supported by NSF award MCB-1406971.