Current Research
Under the auspices of the National Science Foundation, our lab works on the development of a transposon-based functional genomics resource in maize and on the effect of genome structure variation on homologous meiotic recombination.
New reverse genetics resources for maize
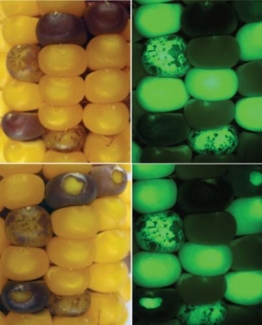
Our lab is presently working on a project funded by the NSF-Plant Genome Program to develop a new reverse genetics resource for the maize community based on the classical McClintock transposable elements (TE) Ac and Ds. Mutations induced by TE insertions help researchers to elucidate gene function. Sequence-indexed insertions (i.e., those in which the host DNA adjacent to the insertion is known) are valuable resources in organisms with a sequenced genome and are deemed essential to fully exploit the recently released maize genome sequence. We are taking advantage of next-generation sequencing technology to develop a rapid and cost- effective method for generating, sequencing, and indexing TE insertions in maize. Specifically, we are sequencing an existing collection of insertions of the native TE Activator (Ac) and new collections of insertions of Ac-derived elements called Ds* that carry the jellyfish green fluorescent protein (GFP) to facilitate tracking their movements in the genome (Figure 1). So far, we have sequence-indexed over 250 Ac and Ds insertions in the maize genome and added them to our new website (acdsinsertions.org), where they will be cross-referenced to stocks available this fall from the maize stock center. The development of this web-searchable database is the result of a collaboration with Drs. Charles Du and Wenwei Xiong at Montclair State University.
In Figure 1, the four views are from the same ear sector, under natural light on the left and under blue light to detect green fluorescence on the right. The purple kernels in the bottom have been cut to expose the green fluorescent endosperm. All green fluorescent kernels carry the transposon Ds*(GFP), the spotted kernels at the original location and the purple kernels at new locations in the genome.
As part of this project, we are creating a genome-wide gene knockout resource for the community that will consist of about 120 roughly equidistant Ds* launching platforms carrying GFP or other easily scored markers. This resource will allow simple visual selection of element transposition from any region of the genome and will enable researchers to generate regional gene knock-out collections because Ac and Ds tend to transpose to nearby chromosomal sites.
Gene conversion at the bronze (bz) locus
Gene conversion involves a nonreciprocal transfer of DNA between two homologous sequences and leads to the replacement of one sequence by its homolog. Allelic gene conversion events most likely correspond to the early recombination nodules detected in synaptonemal complexes and are generated by a distinct recombination pathway from crossovers. In maize, intragenic recombinants (IGRs) carrying a parental or noncrossover arrangement of flanking markers generally arise from conversion events. The stretch of DNA that is transferred during a gene conversion event, called the conversion tract, can vary in organisms like yeast from a few hundred bases to more than 12 kb, but its length has rarely been measured in plants. In most maize studies, IGRs are recovered from heterozygotes between polymorphic alleles that differ at many sites. Because noncrossover IGRs from such heterozygotes are rare, only a few conversion tracts have been measured to date and their minimum length has ranged from 0.5 to 3 kb. We have found that, surprisingly, noncrossover convertants are the predominant IGRs in dimorphic heterozygotes between alleles differing only at the two markers between which recombination is being measured.
Estimating conversion tract lengths in dimorphic heterozygotes, a basically homozygous configuration, is a difficult problem. We have developed a method to measure the minimum- maximum lengths of these conversion tracts that utilizes unselected Ac excision footprints at various distances from the bz gene. We have established by this approach that conversion tracts may be considerably shorter in dimorphic heterozygotes than in polymorphic heterozygotes.