McKim Lab
Research Overview
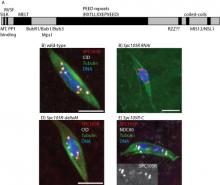
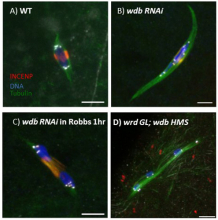
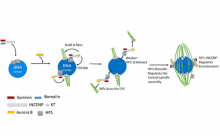
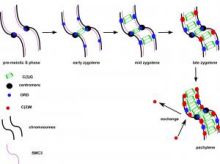
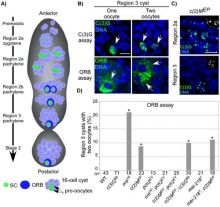
Genome integrity depends on the fidelity of chromosome segregation during mitosis and meiosis. Defects in these processes results in an abnormal number of chromosomes, or aneuploidy, which is usually catastrophic and is the leading cause of infertility in women and the cause of disorders such ‘as Down’s syndrome and is usually associated with tumor cell development. Research in the McKim laboratory is directed at understanding the mechanism of chromosome segregation during meiosis in the fruit fly Drosophila melanogaster. Our experiments include several innovative approaches that combine sophisticated genetics and high-resolution immunofluorescence microscopy to image the structure and behavior of meiotic proteins, chromosomes and spindles in oocytes.
Using molecular genetics, cell biology and genomic approaches, the McKim lab identifies and characterizes genes that are important for all aspects of the meiotic pathway including: i) the assembly of meiotic chromatin, ii) the pairing of homologous chromosomes, iii) recombination between chromosomes, iv) the assembly of a meiotic spindle and v) the orientation and segregation of homologous chromosome. Many of the important genes in this process are also involved in maintaining genome integrity and the fidelity of chromosome division in other cell types. Therefore, these studies also provide insights into the factors affecting genome stability in mitotic cells and the development of cancer cells.
Announcements
-
The latest
Jessica Fellmeth, post-doc, has left the lab to start her own faculty position at Millersville University. Congratulations!
Contact Information
Waksman Institute of Microbiology
190 Frelinghuysen Road
McKim Lab, Office 206
Piscataway, NJ 08854
United States
Selected Publications
Complete list of publications: [Pubmed]
Current Lab Members
Principal Investigator
-
Kim McKim is a Principal Investigator at the Waksman Institute and Professor of Genetics in the School of Arts and Science. Professor McKim uses the fruit fly Drosophila melanogaster to study the fundamental mechanisms of heredity. His lab uses molecular genetics, cell biology and genomic approaches to identify and characterize genes that are important for reproductive fitness, including the segregation of chromosomes during meiosis and other processes that lead to the the generation of a healthy oocyte.
Complete list of publications here
Undergraduate Students
Alumni
Former Graduate Students
- Lin Ing Wang
Former Undergraduate Students
- Jay Joshi
- Tyler Defosse